Who's onlineThere are currently 0 users and 4 guests online.
User loginBook navigationNavigationLive Traffic MapNew Publications
|
Fig. 6.12: Comparison of results of predicting values of Pb (ppm) using ordinary and regression-kriging.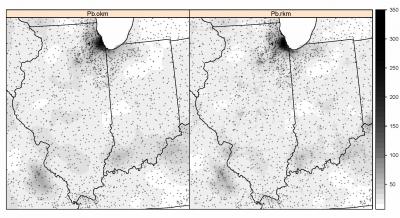
library(maptools)
library(rgdal)
library(RSAGA)
library(gstat)
library(spatstat)
# NGS point data:
load(url("http://spatial-analyst.net/book/system/files/NGS8HMC.Rdata"))
writeOGR(subset(ngs.aea, !is.na(ngs.aea@data[,"PB_ICP40"]))["PB_ICP40"], "PB_ICP40.shp", "PB_ICP40", "ESRI Shapefile")
# USA borders:
load(url("http://spatial-analyst.net/book/system/files/usa_borders.Rdata"))
# Download and extract grids:
download.file("http://spatial-analyst.net/book/system/files/usgrids5km.zip", destfile=paste(getwd(),
"usgrids5km.zip", sep="/"))
grid.list <- c("dairp.asc", "dmino.asc", "dquksig.asc", "dTRI.asc", "gcarb.asc", "geomap.asc",
"globedem.asc", "minotype.asc", "nlights03.asc", "sdroads.asc", "twi.asc", "vsky.asc", "winde.asc", "glwd31.asc")
for(j in grid.list){
fname <- zip.file.extract(file=j, zipname="usgrids5km.zip")
file.copy(fname, paste("./", j, sep=""), overwrite=TRUE)
}
cell.size <- 5000
# ------------------------------------------------------------
# Optional: comparison of OK and RK for a small subset
# ------------------------------------------------------------
# subset the original predictors:
grid.list.s <- c("dairp.asc", "dTRI.asc", "nlights03.asc", "sdroads.asc", "geomap.asc")
rsaga.esri.to.sgrd(in.grids=grid.list.s, out.sgrds=set.file.extension(grid.list.s, ".sgrd"), in.path=getwd())
for(i in 1:length(grid.list.s)) {
# first, create a new grid:
rsaga.geoprocessor(lib="grid_tools", module=23, param=list(GRID="tmp2.sgrd", M_EXTENT=1,
XMIN=360000, YMIN=1555000, XMAX=985000, YMAX=2210000, CELLSIZE=5000))
# 0.01 decimal places:
rsaga.geoprocessor("grid_calculus", module=1, param=list(INPUT="tmp2.sgrd",
RESULT=paste("m_", set.file.extension(grid.list.s[i], ".sgrd"), sep=""), FORMUL="a/100")) # 0.01 decimal places
# now, resample all grids:
rsaga.geoprocessor(lib="grid_tools", module=0, param=list(INPUT=set.file.extension(grid.list.s[i], ".sgrd"),
GRID=paste("m_", set.file.extension(grid.list.s[i], ".sgrd"), sep=""),
GRID_GRID=paste("m_", set.file.extension(grid.list.s[i], ".sgrd"), sep=""),
METHOD=2, KEEP_TYPE=FALSE, SCALE_DOWN_METHOD=0))
}
rsaga.sgrd.to.esri(in.sgrds=paste("m_", set.file.extension(grid.list.s, ".sgrd"), sep=""),
out.grids=paste("m_", set.file.extension(grid.list.s, ".asc"), sep=""), out.path=getwd(), pre=3)
# read maps to R:
gridmaps.s <- readGDAL(paste("m_", set.file.extension(grid.list.s[1], ".asc"), sep=""))
for(i in 2:length(grid.list.s)) {
gridmaps.s@data[i] <- readGDAL(paste("m_", set.file.extension(grid.list.s[i], ".asc"), sep=""))$band1
}
names(gridmaps.s) <- sub(".asc", "", grid.list.s)
str(gridmaps.s@data)
# subset the point data:
rsaga.geoprocessor(lib="shapes_tools", module=14, param=list(SHAPES="PB_ICP40.shp", CUT="m_PB_ICP40.shp",
METHOD=0, TARGET=0, CUT_AX=360000, CUT_BX=985000, CUT_AY=1555000, CUT_BY=2210000))
# prepare the point data in geodata format:
m_PB <- readOGR("m_PB_ICP40.shp", "m_PB_ICP40")
# fix duplicate points:
m_PB <- remove.duplicates(m_PB)
m_PB.ov <- overlay(gridmaps.s, m_PB)
library(geoR)
Pb.geo <- as.geodata(m_PB["PB_ICP40"])
# subset for variogram fitting:
sel <- runif(length(m_PB@data[[1]]))<0.3
Pb.geo1 <- as.geodata(m_PB[sel, "PB_ICP40"])
str(Pb.geo1[[2]])
# prediction locations:
locs <- pred_grid(c(gridmaps.s@bbox[1,1]+cell.size/2, gridmaps.s@bbox[1,2]-cell.size/2),
c(gridmaps.s@bbox[2,1]+cell.size/2, gridmaps.s@bbox[2,2]-cell.size/2), by=cell.size)
# Pb.geo$covariate <- m_PB.ov@data[,sub(".asc", "", grid.list.s)]
Pb.geo1$covariate <- m_PB.ov@data[sel, sub(".asc", "", grid.list.s)]
# prepare the covariates:
locs.sp <- locs
coordinates(locs.sp) <-~Var1+Var2
gridmaps.gr <- overlay(gridmaps.s, locs.sp)
# Ordinary kriging
# fit variogram using likfit:
# Pb.svar <- variog(Pb.geo, lambda=0, max.dist=150000, messages=FALSE)
Pb.vgm <- likfit(Pb.geo1, lambda=0, messages=FALSE,
ini=c(var(log1p(Pb.geo$data)), 50000), cov.model="exponential")
Pb.vgm
Pb.ok <- krige.conv(Pb.geo1, locations=locs, krige=krige.control(obj.m=Pb.vgm))
# Regression-kriging:
# fit variogram model:
Pb.rvgm <- likfit(Pb.geo1, lambda=0, trend=~dairp+dTRI+nlights03+sdroads, messages=FALSE,
ini=c(var(log1p(Pb.geo$data))/5, 25000), cov.model="exponential")
Pb.rvgm
# define the geostatistical model:
KC <- krige.control(trend.d=~dairp+dTRI+nlights03+sdroads,
trend.l=~gridmaps.gr$dairp+gridmaps.gr$dTRI+gridmaps.gr$nlights03+gridmaps.gr$sdroads, obj.m=Pb.rvgm)
# run predictions (external trend kriging):
Pb.rk <- krige.conv(Pb.geo1, locations=locs, krige=KC)
# sp plot:
locs.geo <- data.frame(X=locs.sp@coords[,1], Y=locs.sp@coords[,2], Pb.rk=Pb.rk[[1]],
Pb.ok=Pb.ok[[1]], Pb.rkvar=Pb.rk[[2]], Pb.okvar=Pb.ok[[2]])
coordinates(locs.geo) <-~X+Y
gridded(locs.geo) <- TRUE
# mask out water bodies:
mask.s <- as.vector(t(as.im(gridmaps.s["geomap"])$v)) # flip pixels up-side down
locs.geo$Pb.ok <- ifelse(is.na(mask.s), NA, locs.geo$Pb.ok)
locs.geo$Pb.rk <- ifelse(is.na(mask.s), NA, locs.geo$Pb.rk)
# plot the two maps next to each other:
spplot(locs.geo[c("Pb.ok", "Pb.rk")], col.regions=grey(rev(seq(0,1,0.025)^2.5)), at=seq(7,350,l=40),
sp.layout=list(list("sp.points", m_PB, pch="+", col="black"),
list("sp.lines", USA.borders, col="black"))) # scales=list(draw=TRUE, cex=0.7),
# prediction error:
spplot(locs.geo[c("Pb.okvar", "Pb.rkvar")], col.regions=grey(rev(seq(0,1,0.025)^4)),
at=seq(0,40000,l=40), scales=list(draw=TRUE, cex=0.7),
sp.layout=list("sp.points", m_PB, pch="+", col="black"))
|
Testimonials"I've been looking at your book - I think it provides nice introductions to some important topics. I'm impressed that it's free!" Poll |



Recent comments
6 years 50 weeks ago
7 years 15 weeks ago
7 years 24 weeks ago
7 years 36 weeks ago